Networks Can Be Used to Analyze the Genomes of Humans and Model Organisms
Networks Can Be Used to Analyze the Genomes of Humans and Model Organisms
Soon, sequencing one’s genome may become as commonplace as getting an X-ray. Consequently, personal genomes will increasingly serve as the lenses through which the public views biology. Addressing this, the focus of the Gerstein Lab is interpreting personal genomes, particularly in relation to disorders such as cancer. This endeavor has a number of related aspects.
The Analysis of Diverse Networks
Networks are a way of tying together much of our research. Network representations can be applied consistently to many different types of biological data; thus, we have developed tools to build and analyze regulatory networks, protein-protein interactions and metabolic pathways, identifying key nodes such as hubs and bottlenecks.
Moreover, because they are a generic and flexible representation, networks provide an ideal framework for data integration. We have integrated networks with dynamic gene-expression data (identifying transient hubs), 3D-protein structures, and even satellite imagery.
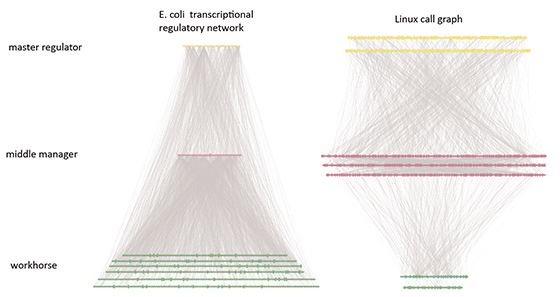
Finally, as people have more intuition for commonplace networks, such as those in social and computer systems, we have found cross-disciplinary comparisons helpful, elucidating system-level properties of biological networks, such as the association of greater connectivity with more evolutionary constraint.